CAVE: Connectome Annotation Versioning Engine
![]()


Overview
In connectomics we aim to map and understand the intricate networks of the brain at the cellular level. With advancements in volume electron microscopy (EM) and computational technologies, we are now able to capture and reconstruct the neural circuitry in unprecedented detail. However, this explosion of data presents new challenges: how do we manage, proofread, and analyze these vast datasets efficiently and collaboratively?
The Connectome Annotation Versioning Engine (CAVE) is a computational infrastructure designed to address some of these challenges. It enables community proofreading and immediate and reproducible connectome analysis of up to petascale datasets—approximately 1 cubic millimeter in size, equivalent to about 1 petabyte of raw imagery. CAVE is a collection of web-services, typically deployed in the cloud in order to make the data available to anyone anywhere in the world.
As such, it has facilitated reconstruction of the world’s largest datasets including the MICrONs project, Flywire, FANC, and H01 through the collaborative effort of hundreds of scientists, citizen scientists and machine learning algorithms.
CAVE was developed initially through collaboration between Sebastian Seung’s laboratory at Princeton University, and the Electron Microscopy Connectomics project at the Allen Institute for Brain Science.
Why CAVE?
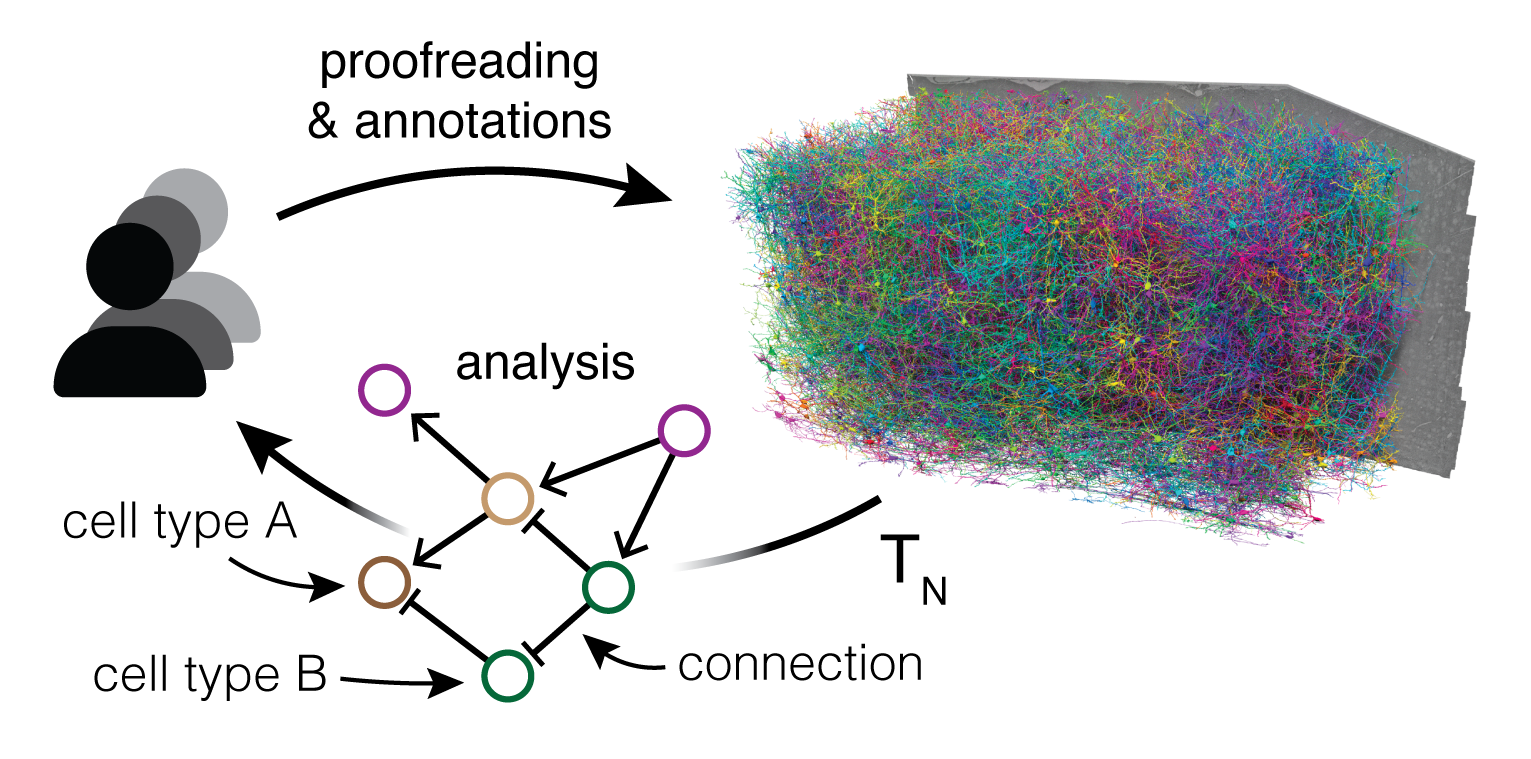
Collaborative Proofreading and Annotation
Proofreading and annotating large-scale connectomic datasets is a time-consuming process, often involving years of manual work. Existing tools limited collaboration, were not able to efficiently scale to the petabyte range, or were not equipped to handle continuous updates. CAVE introduced a distributed proofreading infrastructure, allowing for concurrent annotation and correction of errors in automated segmentation. This means that multiple users can work together seamlessly, making the process faster and more efficient.
Immediate Analysis
One of the unique challenges in connectomics is the need for immediate analysis post-proofreading. In CAVE, annotations are bound to segment IDs at specific points in time, a process we call “materialization.”
This allows for fast queries and analysis immediately after edits, without delays. Whether you’re interested in the current state of the data or a snapshot from a specific time point, CAVE delivers accurate, on-demand results.
Extensible and Scalable
CAVE is flexible in how annotations are defined. It supports schematized, extensible annotations, making it easy for researchers to design and implement novel annotation types. Users can create new types of annotations and create different tables that utilize the same types of annotations. This allows communities to more naturally collaborate and compare different approaches to problems such as cell-typing in a dataset. It also allows the number of use cases the platform can cover to grow quickly. Annotating a new sub-cellular organelle type, or integrating new observations about sub-cellular targetting patterns of axons is easy to extend. Additionally, CAVE’s infrastructure is built to scale, efficiently handling petascale datasets and supporting the continuous versioning of large reconstructions.
CAVE is organized as a loosely coupled set of micro-services, each of which has an open API which can be used by services either inside or outside CAVE. This means that CAVE can easily be built on top of to create new services or libraries to benefit communities. Examples include NeuView, FAFBSEG, braincircuits.io, NAVIS and CODEX.
Website organization
Whether you’re a neuroscientist looking to explore and analyze connectomic datasets, a developer interested in understanding or contributing to CAVE’s underlying services, or someone tasked with deploying CAVE as a cloud service, this website is your starting point. We have organized the website into sections that are focused on these different audiences.
- Users: Discover how to access and utilize CAVE for your research.
- Developers (COMING SOON): Dive into the technical details of CAVE’s services, access the codebase, and learn how to contribute.
- Deployment: Find guidance on how to deploy CAVE as a cloud service, ensuring scalable and efficient access.
Read the papers
The most comprehensive description of CAVE can be found in the CAVE paper. Earlier descriptions of the proofreading system can be found in the original FlyWire manuscript and was first developed and used to do proofreading and analysis in the MICrONS phase 1 dataset.
Publicly available datasets and communities using CAVE

|
MICrONS datasets. The MICrONS consortium created multiple structure-function datasets from mouse visual cortext up to a 1 mm3. See also the MICrONS explorer website. |

|
FlyWire. The FlyWire consortium proofread the first whole brain connectome from the fruit fly. The dataset is now publicly available. See also the FlyWire website. |